##################################### # From Scratch Power Analysis Monte Carlo: Survey ###################################### # Clear Environment rm (list= ls ())library (EnvStats, quietly = TRUE , warn.conflicts= FALSE )library (truncnorm, quietly = TRUE , warn.conflicts= FALSE )library (ggplot2, quietly = TRUE , warn.conflicts= FALSE )library (haven, quietly = TRUE , warn.conflicts= FALSE )
Warning: package 'haven' was built under R version 4.3.3
library (readr, quietly = TRUE , warn.conflicts= FALSE )library (tidyverse, quietly = TRUE , warn.conflicts= FALSE )
Warning: package 'tidyverse' was built under R version 4.3.3
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ stringr 1.5.1
✔ forcats 1.0.0 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library (MASS, quietly = TRUE , warn.conflicts= FALSE )library (ggplot2, quietly = TRUE , warn.conflicts= FALSE )library (dplyr, quietly = TRUE , warn.conflicts= FALSE )library (tidyr, quietly = TRUE , warn.conflicts= FALSE )library (faux, quietly = TRUE , warn.conflicts= FALSE )
************
Welcome to faux. For support and examples visit:
https://debruine.github.io/faux/
- Get and set global package options with: faux_options()
************
<- read_dta ("DTA_Files/survey_power_analysis.dta" )# mcdata <- read_dta("update_power_analysis_results_mediation.dta") <- mcdata %>% group_by (sample_size, beta1, beta2, beta3,beta4,beta5,beta6,beta7) %>% :: summarize (H1_mig = sum (H1_agg_mig),H1_imp = sum (H1_agg_imp),H1_auto = sum (H1_agg_auto),H1_off = sum (H1_agg_off),H2_mig = sum (H2_agg_mig),H2_imp = sum (H2_agg_imp),H2_auto = sum (H2_agg_auto),H2_off = sum (H2_agg_off),H3_mig = sum (H3_agg_mig),H3_imp = sum (H3_agg_imp),H3_auto = sum (H3_agg_auto),H3_off = sum (H3_agg_off),H4_mig = sum (H4_agg_mig),H4_imp = sum (H4_agg_imp),H4_auto = sum (H4_agg_auto),H4_off = sum (H4_agg_off),H5_imp = sum (H5_agg_imp),H5_auto = sum (H5_agg_auto),H5_off = sum (H5_agg_off),H6_mig = sum (H6_agg_mig),H6_imp = sum (H6_agg_imp),H6_auto = sum (H6_agg_auto),H6_off = sum (H6_agg_off),H7_mig = sum (H7_agg_mig),H7_imp = sum (H7_agg_imp),H7_auto = sum (H7_agg_auto),H7_off = sum (H7_agg_off)
`summarise()` has grouped output by 'sample_size', 'beta1', 'beta2', 'beta3',
'beta4', 'beta5', 'beta6'. You can override using the `.groups` argument.
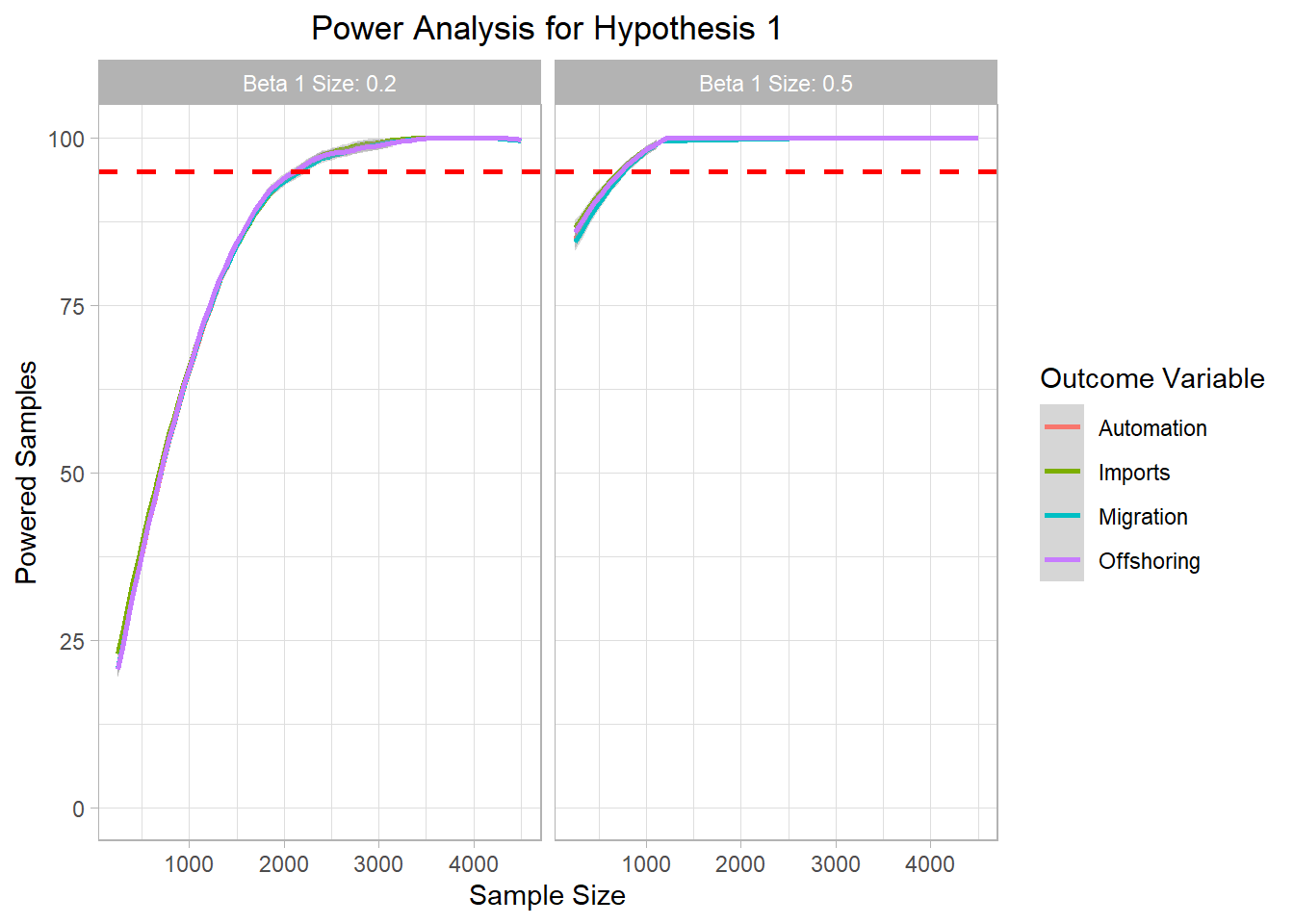
$ beta1_factor <- as.character (mc_data_sum$ beta1)$ beta2_factor <- as.character (mc_data_sum$ beta2)$ beta3_factor <- as.character (mc_data_sum$ beta3)$ beta4_factor <- as.character (mc_data_sum$ beta4)$ beta5_factor <- as.character (mc_data_sum$ beta5)$ beta6_factor <- as.character (mc_data_sum$ beta6)$ beta7_factor <- as.character (mc_data_sum$ beta7)$ beta1_factor <- paste ("Beta 1 Size:" , mc_data_sum$ beta1_factor, sep = " " )$ beta2_factor <- paste ("Beta 2 Size:" , mc_data_sum$ beta2_factor, sep = " " )$ beta3_factor <- paste ("Beta 3 Size:" , mc_data_sum$ beta3_factor, sep = " " )$ beta4_factor <- paste ("Beta 4 Size:" , mc_data_sum$ beta4_factor, sep = " " )$ beta5_factor <- paste ("Beta 5 Size:" , mc_data_sum$ beta5_factor, sep = " " )$ beta6_factor <- paste ("Beta 6 Size:" , mc_data_sum$ beta6_factor, sep = " " )$ beta7_factor <- paste ("Beta 7 Size:" , mc_data_sum$ beta7_factor, sep = " " )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H1_mig, H1_imp, H1_auto, H1_off), names_to = "H1" ,values_to = "H1_agg" )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H2_mig, H2_imp, H2_auto, H2_off), names_to = "H2" ,values_to = "H2_agg" )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H3_mig, H3_imp, H3_auto, H3_off), names_to = "H3" ,values_to = "H3_agg" )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H4_mig, H4_imp, H4_auto, H4_off), names_to = "H4" ,values_to = "H4_agg" )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H5_imp, H5_auto, H5_off), names_to = "H5" ,values_to = "H5_agg" )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H6_mig, H6_imp, H6_auto, H6_off), names_to = "H6" ,values_to = "H6_agg" )<- mc_data_sum %>% tidyr:: pivot_longer (cols= c (H7_mig, H7_imp, H7_auto, H7_off), names_to = "H7" ,values_to = "H7_agg" )$ H1 <- replace (mc_data_H1$ H1, mc_data_H1$ H1== "H1_auto" ,"Automation" )$ H1 <- replace (mc_data_H1$ H1, mc_data_H1$ H1== "H1_imp" ,"Imports" )$ H1 <- replace (mc_data_H1$ H1, mc_data_H1$ H1== "H1_off" ,"Offshoring" )$ H1 <- replace (mc_data_H1$ H1, mc_data_H1$ H1== "H1_mig" ,"Migration" )$ H2 <- replace (mc_data_H2$ H2, mc_data_H2$ H2== "H2_auto" ,"Automation" )$ H2 <- replace (mc_data_H2$ H2, mc_data_H2$ H2== "H2_imp" ,"Imports" )$ H2 <- replace (mc_data_H2$ H2, mc_data_H2$ H2== "H2_off" ,"Offshoring" )$ H2 <- replace (mc_data_H2$ H2, mc_data_H2$ H2== "H2_mig" ,"Migration" )$ H3 <- replace (mc_data_H3$ H3, mc_data_H3$ H3== "H3_auto" ,"Automation" )$ H3 <- replace (mc_data_H3$ H3, mc_data_H3$ H3== "H3_imp" ,"Imports" )$ H3 <- replace (mc_data_H3$ H3, mc_data_H3$ H3== "H3_off" ,"Offshoring" )$ H3 <- replace (mc_data_H3$ H3, mc_data_H3$ H3== "H3_mig" ,"Migration" )$ H4 <- replace (mc_data_H4$ H4, mc_data_H4$ H4== "H4_auto" ,"Automation" )$ H4 <- replace (mc_data_H4$ H4, mc_data_H4$ H4== "H4_imp" ,"Imports" )$ H4 <- replace (mc_data_H4$ H4, mc_data_H4$ H4== "H4_off" ,"Offshoring" )$ H4 <- replace (mc_data_H4$ H4, mc_data_H4$ H4== "H4_mig" ,"Migration" )$ H5 <- replace (mc_data_H5$ H5, mc_data_H5$ H5== "H5_auto" ,"Automation" )$ H5 <- replace (mc_data_H5$ H5, mc_data_H5$ H5== "H5_imp" ,"Imports" )$ H5 <- replace (mc_data_H5$ H5, mc_data_H5$ H5== "H5_off" ,"Offshoring" )$ H5 <- replace (mc_data_H5$ H5, mc_data_H5$ H5== "H5_mig" ,"Migration" )$ H6 <- replace (mc_data_H6$ H6, mc_data_H6$ H6== "H6_auto" ,"Automation" )$ H6 <- replace (mc_data_H6$ H6, mc_data_H6$ H6== "H6_imp" ,"Imports" )$ H6 <- replace (mc_data_H6$ H6, mc_data_H6$ H6== "H6_off" ,"Offshoring" )$ H6 <- replace (mc_data_H6$ H6, mc_data_H6$ H6== "H6_mig" ,"Migration" )$ H7 <- replace (mc_data_H7$ H7, mc_data_H7$ H7== "H7_auto" ,"Automation" )$ H7 <- replace (mc_data_H7$ H7, mc_data_H7$ H7== "H7_imp" ,"Imports" )$ H7 <- replace (mc_data_H7$ H7, mc_data_H7$ H7== "H7_off" ,"Offshoring" )$ H7 <- replace (mc_data_H7$ H7, mc_data_H7$ H7== "H7_mig" ,"Migration" )<- mc_data_H1 %>% ggplot (aes (x= sample_size, y= H1_agg, color= H1))+ geom_smooth ()+ ylim (0 ,100 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 1" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ facet_wrap (~ beta1_factor)+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h1_plot
Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
Warning: Removed 253 rows containing missing values (`geom_smooth()`).
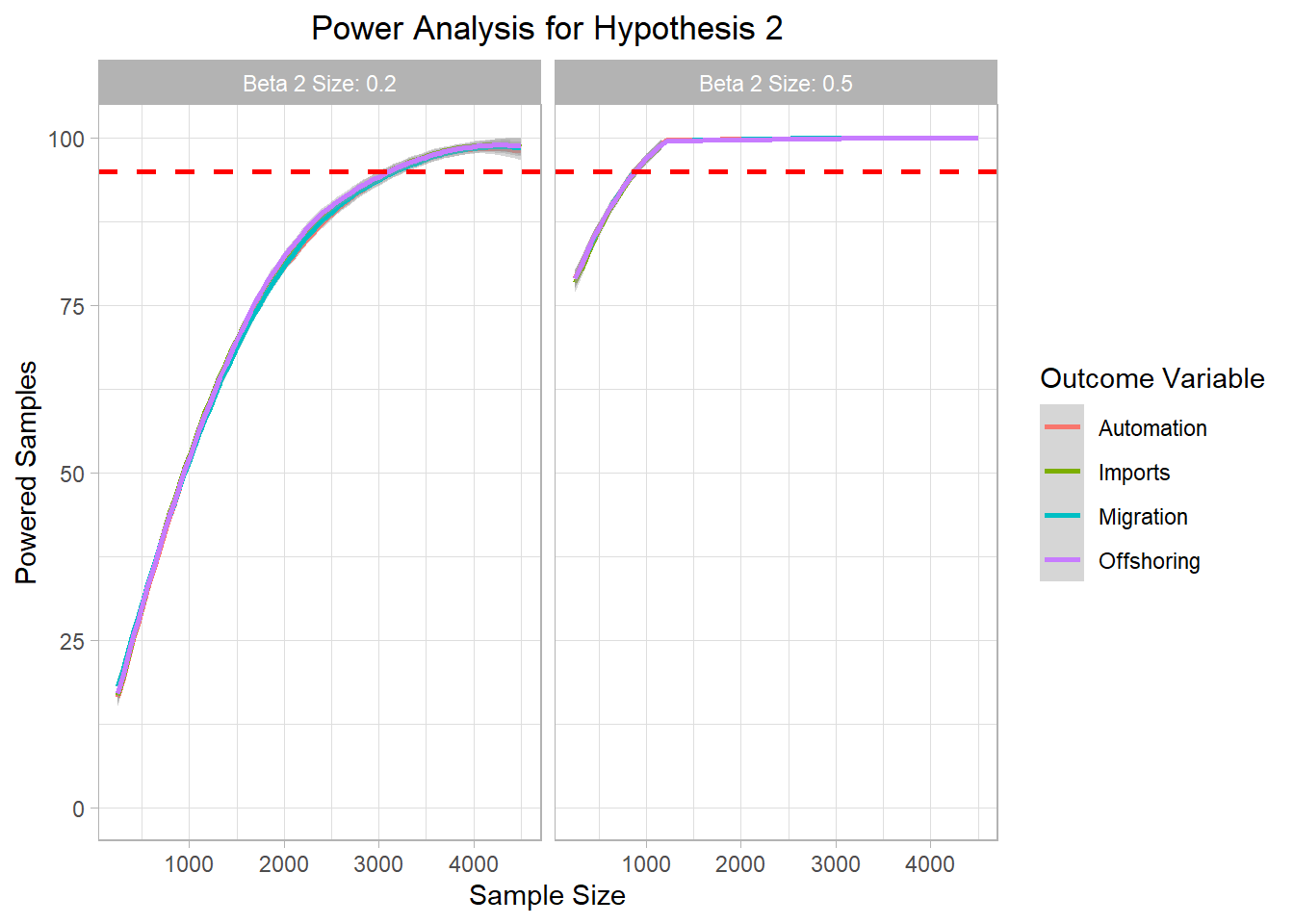
# ggsave("Graphics/survey_h1_plot.png") <- mc_data_H2 %>% ggplot (aes (x= sample_size, y= H2_agg, color= H2))+ geom_smooth ()+ ylim (0 ,100 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 2" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ facet_wrap (~ beta2_factor)+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h2_plot
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
Warning: Removed 212 rows containing missing values (`geom_smooth()`).
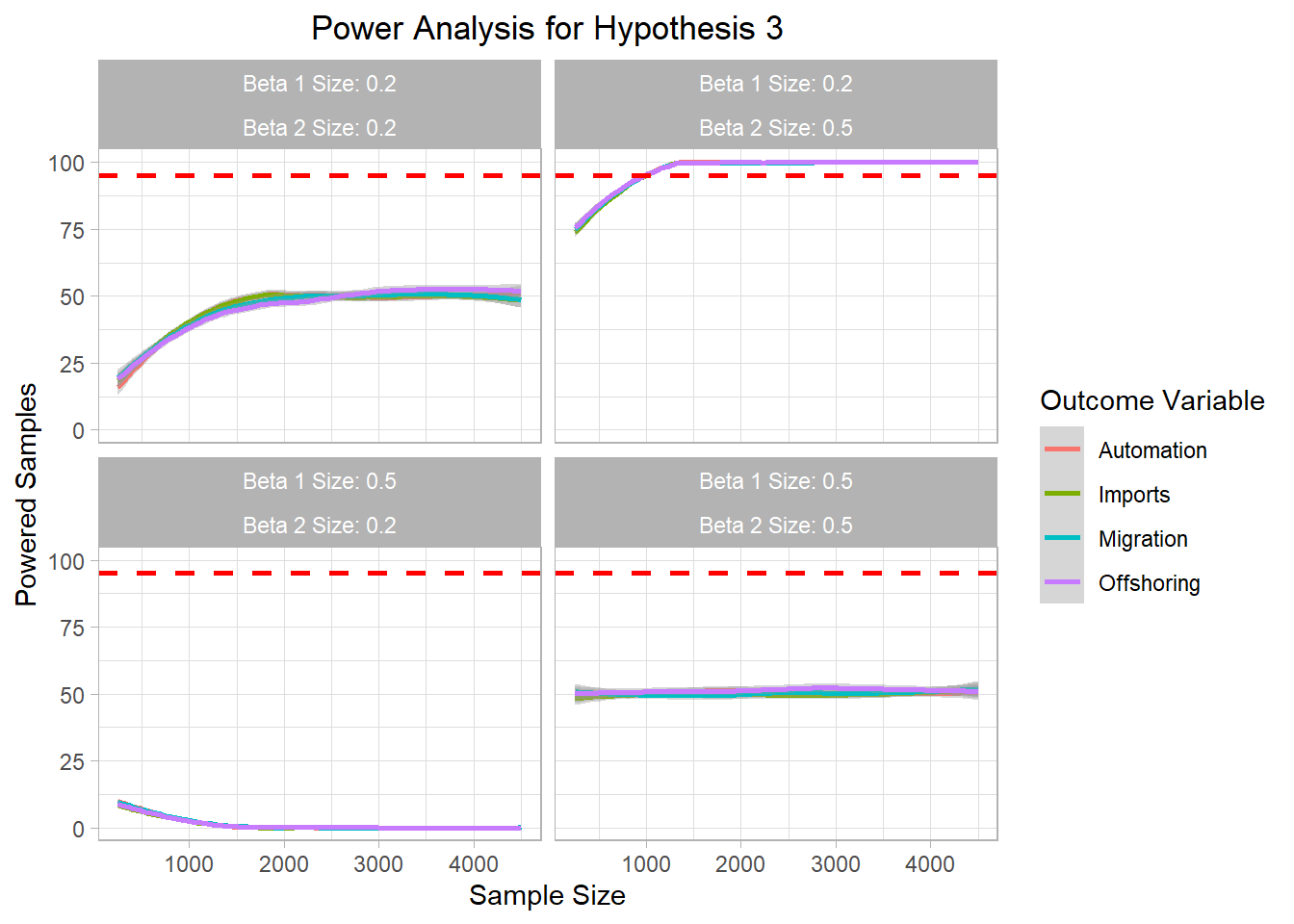
# ggsave("Graphics/survey_h2_plot.png") <- mc_data_H3 %>% ggplot (aes (x= sample_size, y= H3_agg, color= H3))+ geom_smooth ()+ ylim (0 ,100 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 3" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ facet_wrap (~ beta1_factor + beta2_factor)+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h3_plot
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
Warning: Removed 326 rows containing missing values (`geom_smooth()`).
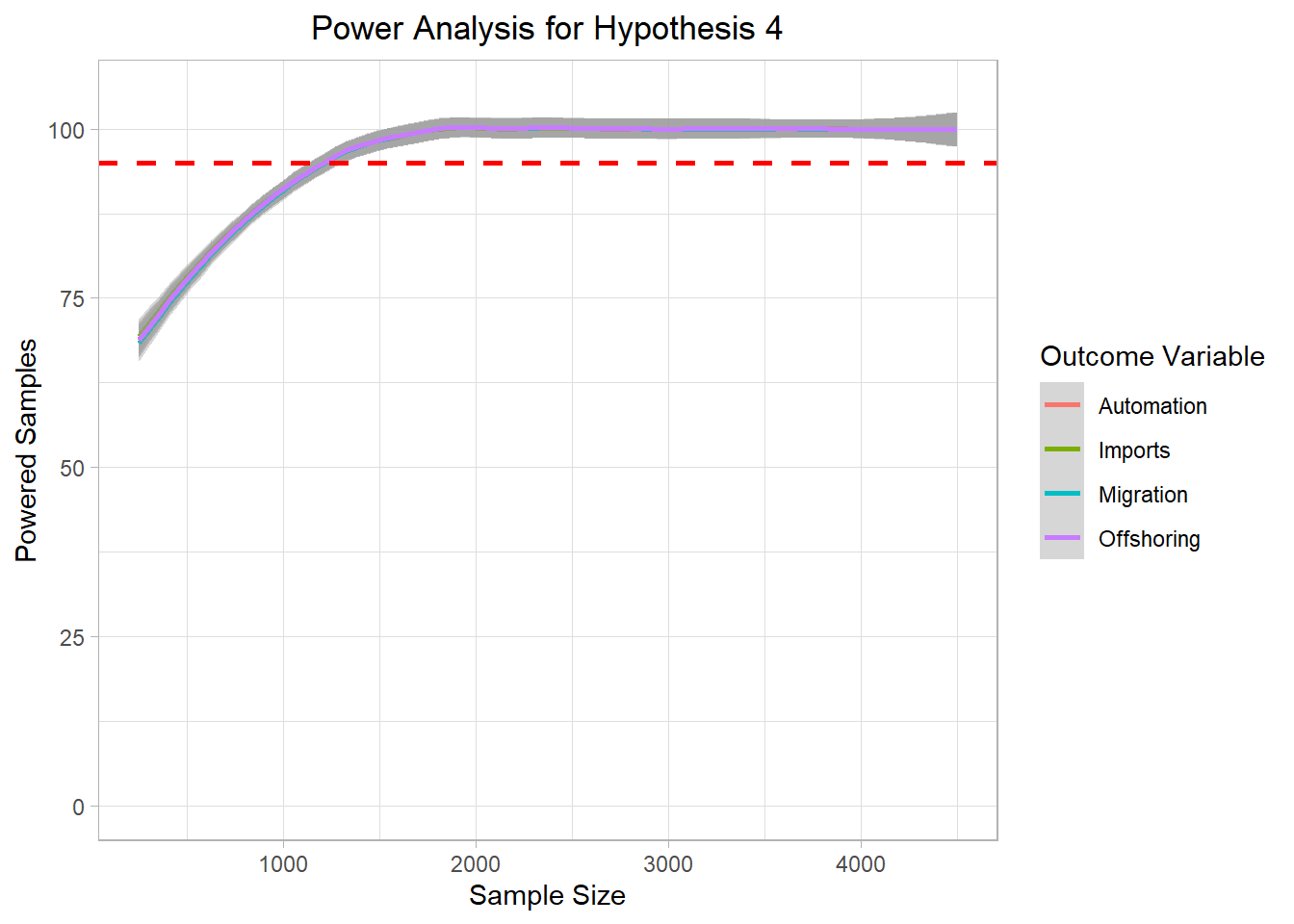
# ggsave("Graphics/survey_h3_plot.png") <- mc_data_H4 %>% ggplot (aes (x= sample_size, y= H4_agg, color= H4))+ geom_smooth ()+ ylim (0 ,105 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 4" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ # facet_wrap(~beta3_factor + beta6_factor + beta2_factor)+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h4_plot
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
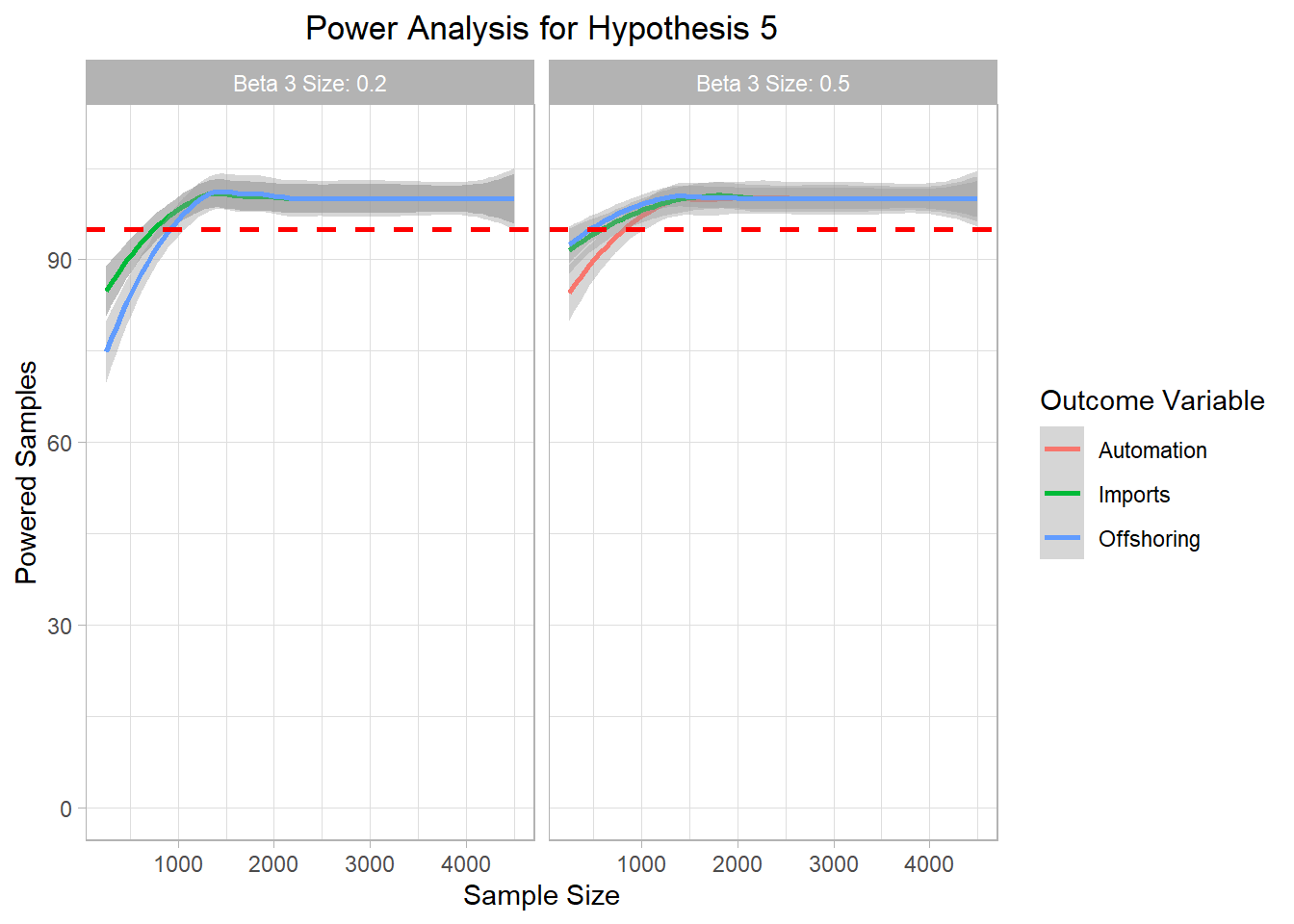
# ggsave("Graphics/survey_h4_plot.png") <- mc_data_H5 %>% ggplot (aes (x= sample_size, y= H5_agg, color= H5))+ geom_smooth ()+ ylim (0 ,110 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 5" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ facet_wrap (~ beta3_factor)+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h5_plot
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
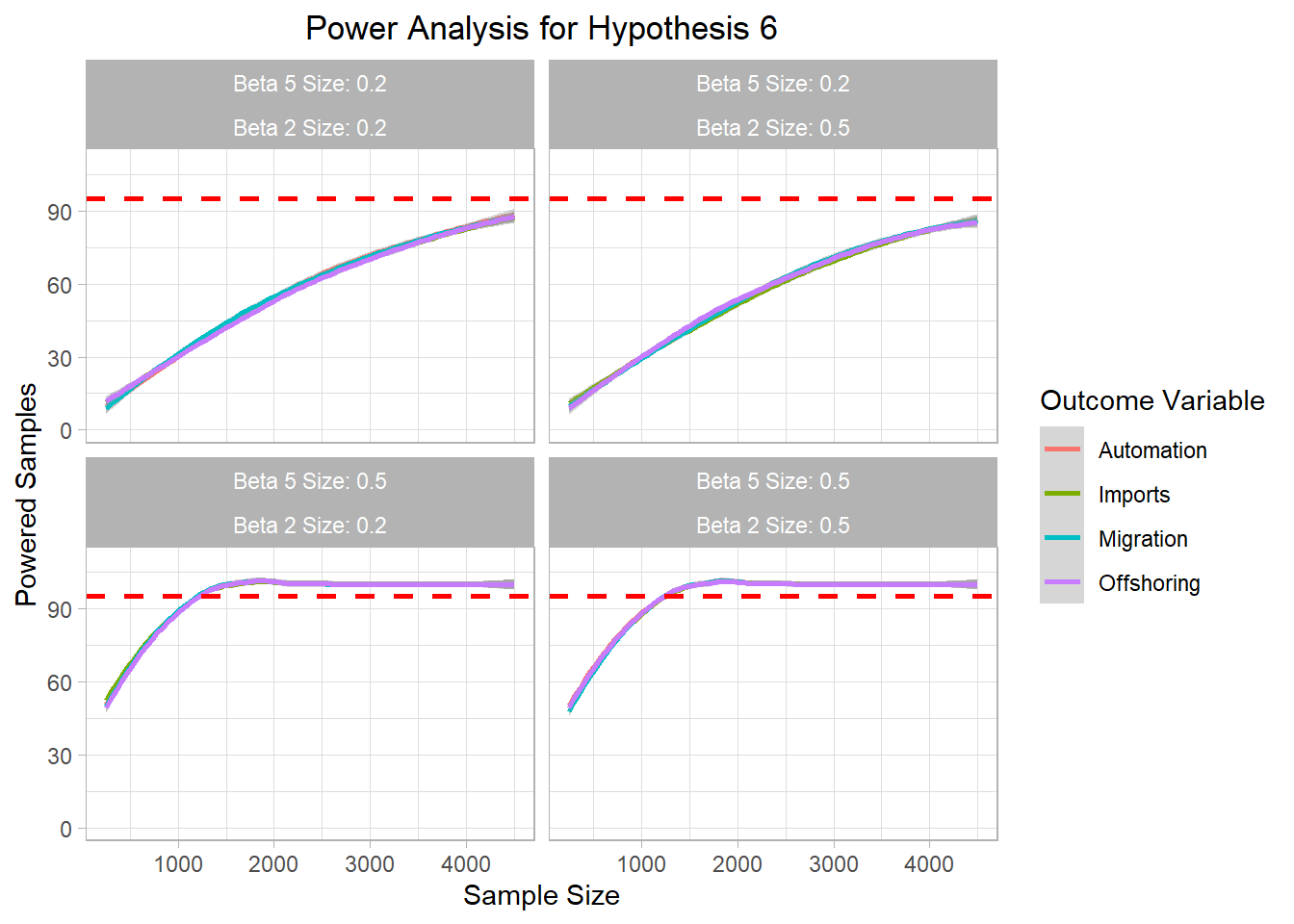
# ggsave("Graphics/survey_h5_plot.png") <- mc_data_H6 %>% ggplot (aes (x= sample_size, y= H6_agg, color= H6))+ geom_smooth ()+ ylim (0 ,110 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 6" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ facet_wrap (~ beta5_factor + beta2_factor)+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h6_plot
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
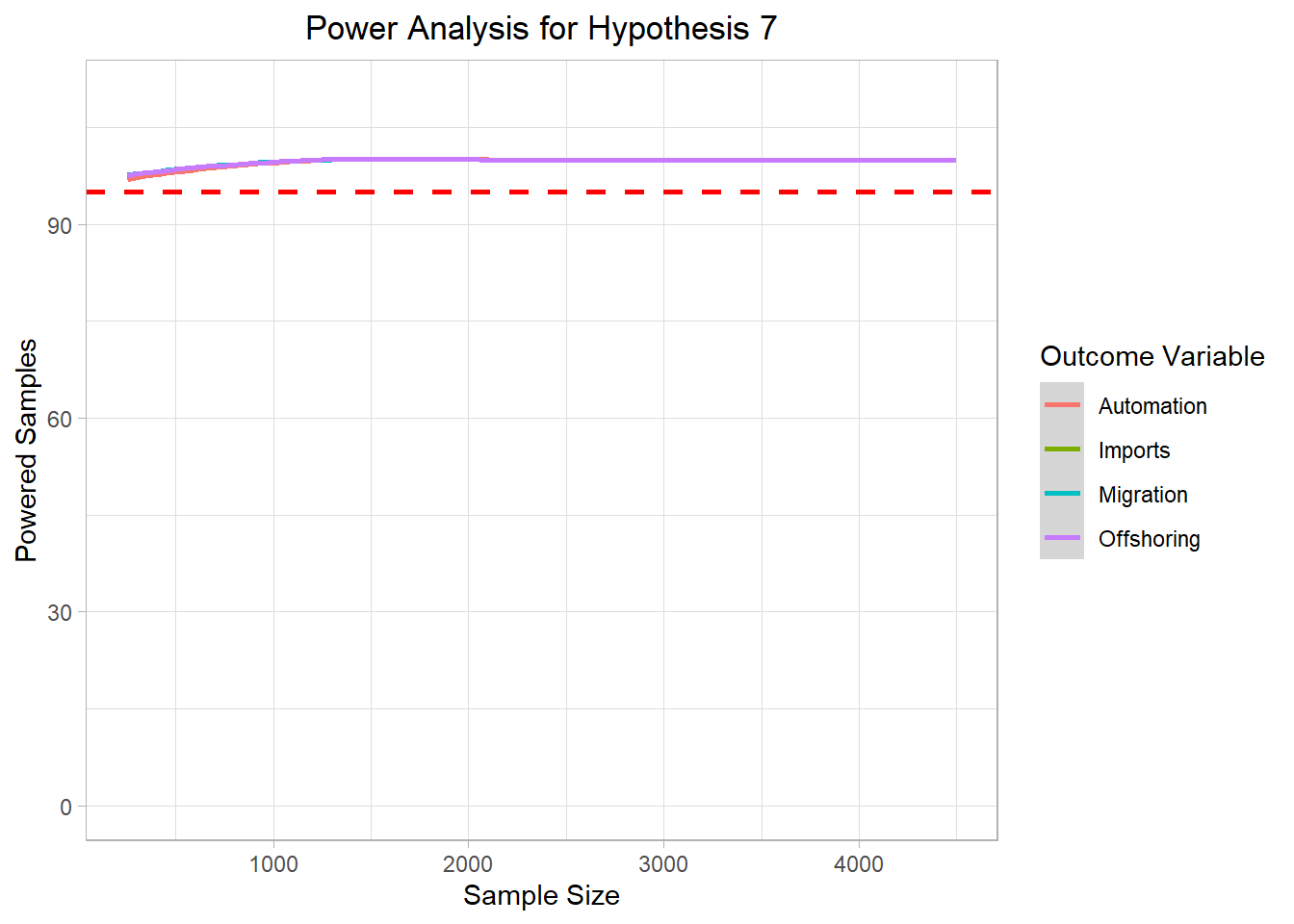
# ggsave("Graphics/survey_h6_plot.png") <- mc_data_H7 %>% ggplot (aes (x= sample_size, y= H7_agg, color= H7))+ geom_smooth ()+ ylim (0 ,110 )+ geom_hline (yintercept= 95 , col= "red" ,lty= "dashed" , size= 1 )+ labs (title= "Power Analysis for Hypothesis 7" ,x= "Sample Size" ,color= "Outcome Variable" ,y= "Powered Samples" )+ theme_light ()+ theme (plot.title = element_text (hjust = 0.5 ));h7_plot
`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
# ggsave("Graphics/survey_h7_plot.png")